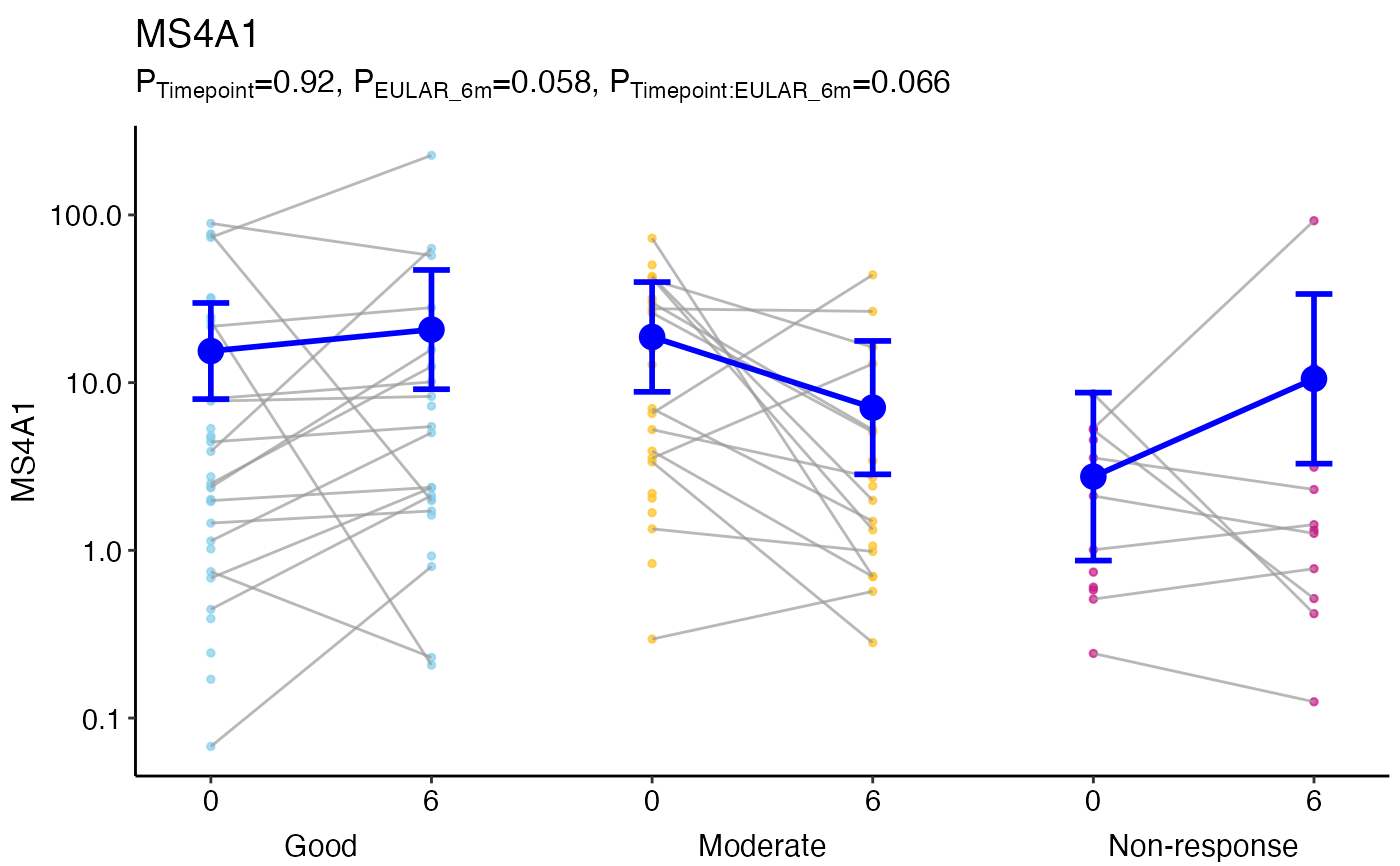
Plot to show differences between groups and over time using ggplot2.
Usage
ggmodelPlot(
object,
geneName = NULL,
x1var = NULL,
x2var = NULL,
x2shift = NULL,
xlab = NULL,
ylab = geneName,
plab = NULL,
title = geneName,
logTransform = is(object, "GlmmSeq"),
shapes = 19,
colours = "grey60",
lineColours = "grey60",
markerSize = 1,
fontSize = 12,
alpha = 0.7,
x2Offset = 5,
addPoints = TRUE,
addModel = TRUE,
modelSize = 4,
modelColours = "blue",
modelLineSize = 1,
modelLineColours = modelColours,
addBox = FALSE,
...
)Arguments
- object
A glmmSeq/lmmSeq object created by
glmmSeq::glmmSeq()orglmmSeq::lmmSeq()- geneName
The gene/row name to be plotted
- x1var
The name of the first (inner) x parameter, typically 'time'. This is anticipated to have different values when matched by ID.
- x2var
The name of an optional second (outer) x parameter, which should be a factor.
- x2shift
Amount to shift along x axis for each level of
x2var. By default the function will arrange each level ofx2varside by side.- xlab
Title for the x axis
- ylab
Title for the y axis
- plab
Optional character vector of labels for p-values. These must align with column names in
object@stats$pvals.- title
Plot title. If NULL gene name is used
- logTransform
Whether to perform a log10 transform on the y axis
- shapes
The marker shapes (default=19)
- colours
The marker colours as vector
- lineColours
The line colours (default='grey60') as vector
- markerSize
Size of markers (default=1)
- fontSize
Plot font size
- alpha
Line and marker opacity (default=0.7)
- x2Offset
Vertical adjustment to secondary x-axis labels (default=5)
- addPoints
Whether to add underlying data points (default=TRUE)
- addModel
Whether to add the fit model with markers (default=TRUE)
- modelSize
Size of model points (default=4)
- modelColours
Colour of model fit markers (default="blue") as vector
- modelLineSize
Size of model points (default=1) as vector
- modelLineColours
Colour of model fit lines
- addBox
Logical whether to add boxplots for mean and IQR
- ...
Other parameters to pass to
ggplot2::theme().
Examples
data(PEAC_minimal_load)
disp <- apply(tpm, 1, function(x){
(var(x, na.rm=TRUE)-mean(x, na.rm=TRUE))/(mean(x, na.rm=TRUE)**2)
})
MS4A1glmm <- glmmSeq(~ Timepoint * EULAR_6m + (1 | PATID),
countdata = tpm['MS4A1', , drop = FALSE],
metadata = metadata,
dispersion = disp,
verbose = FALSE)
ggmodelPlot(object = MS4A1glmm,
geneName = 'MS4A1',
x1var = 'Timepoint',
x2var = 'EULAR_6m',
colours = c('skyblue', 'goldenrod1', 'mediumvioletred'))